Model Lineage Tracking
This notebook’s CI test result for us-west-2 is as follows. CI test results in other regions can be found at the end of the notebook.
Amazon SageMaker ML Lineage Tracking creates and stores information about the steps of a machine learning (ML) workflow from data preparation to model deployment. With the tracking information you can reproduce the workflow steps, track model and dataset lineage, and establish model governance and audit standards.
Tracking entities maintain a representation of all the elements of your end-to-end machine learning workflow. You can use this representation to establish model governance, reproduce your workflow, and maintain a record of your work history. Amazon SageMaker automatically creates tracking entities for trial components and their associated trials and experiments when you create SageMaker jobs such as processing jobs, training jobs, and batch transform jobs.
In this example, we will collect the model lineage data and represent it in a Heterogeneous Graph to maintain the structure, cohesion and trace lineage data back to its original source.
Heterogeneous Graphs
Heterogeneous graphs, or heterographs for short, are graphs that contain different types of nodes and edges. The different types of nodes and edges tend to have different types of attributes that are designed to capture the characteristics of each node and edge type. Within the context of graph neural networks, depending on their complexity, certain node and edge types might need to be modeled with representations that have a different number of dimensions. Source
Import SageMaker Libraries
[ ]:
!pip install -U sagemaker boto3 sagemaker-experiments
[ ]:
import boto3
from botocore.config import Config
import sagemaker
from sagemaker.lineage import context, artifact, association, action
from smexperiments import experiment, trial_component
import itertools
config = Config(retries={"max_attempts": 20, "mode": "adaptive"})
client = boto3.client("sagemaker", config=config)
sagemaker_session = sagemaker.Session(sagemaker_client=client)
BUCKET = sagemaker_session.default_bucket()
REGION = boto3.session.Session().region_name
ACCOUNT_ID = boto3.client("sts").get_caller_identity().get("Account")
lookup_dict = {}
LIMIT = 50
Create the Artifacts Nodes
An artifact is a lineage tracking entity that represents a URI addressable object or data. Some examples are the S3 URI of a dataset and the ECR registry path of an image.
[ ]:
header = "~id,~label,arn:string,sourceuri:string,type:string,creationtime,LastModifiedTime"
counter = itertools.count()
arts = artifact.Artifact.list(sagemaker_session=sagemaker_session)
with open("artifacts-nodes.csv", "w") as f:
f.write(header + "\n")
for index, art in enumerate(arts):
idd = str(next(counter))
label = "artifact"
arn = art.artifact_arn
sourceuri = art.source.source_uri
if art.artifact_type is not None:
dtype = art.artifact_type
else:
dtype = ""
ctime = art.creation_time
mtime = art.last_modified_time
line = [idd, label, arn, sourceuri, dtype, ctime, mtime]
lookup_dict.update({arn: idd})
f.write(",".join(map(str, line)) + "\n")
# Limiting the output to 50 artifacts
# Remove the following limit if exporting data for the first time
if index == LIMIT:
break
Create Trial Components Nodes
A trial component is a stage of a machine learning trial. Includes processing jobs, training jobs, and batch transform jobs. In the next cell, we will export all of the trial component data from the account.
[ ]:
header = "~id,~label,arn:string,sourcearn:string,type:string,creationtime,LastModifiedTime,status:string,message:string"
counter = itertools.count()
tcs = trial_component.TrialComponent.list(sagemaker_boto_client=client)
with open("tcs-nodes.csv", "w") as f:
f.write(header + "\n")
for index, tc in enumerate(tcs):
idd = "t" + str(next(counter))
label = "trial_component"
arn = tc.trial_component_arn
if hasattr(tc, "trial_component_source"):
sourcearn = tc.trial_component_source["SourceArn"]
dtype = tc.trial_component_source["SourceType"]
else:
sourcearn = ""
dtype = ""
ctime = tc.creation_time
mtime = tc.last_modified_time
status = tc.status.primary_status if hasattr(tc, "status") and tc.status != None else ""
message = tc.status.message if hasattr(tc.status, "message") else ""
line = [
idd,
label,
arn,
sourcearn,
dtype,
ctime,
mtime,
status,
str(message).replace("\n", " ").replace(",", "-"),
]
lookup_dict.update({arn: idd})
f.write(",".join(map(str, line)) + "\n")
# Limiting the output to 50 trial components
# Remove the following limit if exporting data for the first time
if index == LIMIT:
break
Create Actions Nodes
Actions represents an action or activity. Generally, an action involves at least one input artifact or output artifact. Some examples are a workflow step and a model deployment.
[ ]:
acts = action.Action.list(sagemaker_session=sagemaker_session)
header = (
"~id,~label,arn:string,sourceuri:string,status:string,type:string,creationtime,LastModifiedTime"
)
counter = itertools.count()
with open("action-nodes.csv", "w") as f:
f.write(header + "\n")
for index, act in enumerate(acts):
idd = "ac" + str(next(counter))
label = "action"
arn = act.action_arn
sourceuri = act.source.source_uri
status = act.status if hasattr(act, "status") and act.status != None else ""
dtype = act.action_type
ctime = act.creation_time
mtime = act.last_modified_time
line = [idd, label, arn, sourceuri, status, dtype, ctime, mtime]
lookup_dict.update({arn: idd})
f.write(",".join(map(str, line)) + "\n")
# Limiting the output to 50 actions
# Remove the following limit if exporting data for the first time
if index == LIMIT:
break
Create Context Nodes
Contexts are logical grouping of other tracking or experiment entities. Conceptually, experiments and trials are contexts. Some examples are an endpoint and a model package
[ ]:
ctxs = context.Context.list(sagemaker_session=sagemaker_session)
header = (
"~id,~label,arn:string,sourceuri:string,status:string,type:string,creationtime,LastModifiedTime"
)
counter = itertools.count()
with open("contexts-nodes.csv", "w") as f:
f.write(header + "\n")
for index, ctx in enumerate(ctxs):
idd = "ctx" + str(next(counter))
label = "context"
arn = ctx.context_arn
sourceuri = ctx.source.source_uri
dtype = ctx.context_type
ctime = ctx.creation_time
mtime = ctx.last_modified_time
line = [idd, label, arn, sourceuri, dtype, ctime, mtime]
lookup_dict.update({arn: idd})
f.write(",".join(map(str, line)) + "\n")
# Limiting the output to 50 contexts
# Remove the following limit if exporting data for the first time
if index == LIMIT:
break
Create Edges from Associations
An association is the relationship that links the source and destination entities. For example, an association between the location of training data and a training job. The exported file will be used as the edges information between the entities in the graph. This step may take a few hours to finish.
[ ]:
header = "~id,~from,~to,~label"
counter = itertools.count()
with open("edges.csv", "w") as f:
f.write(header + "\n")
for key in lookup_dict.keys():
associations = association.Association.list(
destination_arn=key, sagemaker_session=sagemaker_session
)
if associations is not None:
for asso in associations:
if asso.source_arn in lookup_dict:
ct = next(counter)
idd = "e" + str(ct)
fr = lookup_dict[asso.source_arn]
to = lookup_dict[asso.destination_arn]
label = asso.association_type if hasattr(asso, "association_type") else ""
line = [idd, fr, to, label]
f.write(",".join(map(str, line)) + "\n")
Now, all the data needed to build the graph is ready. We will use this data to populate a Neptune cluster with the graph data.
Upload the generated files to S3
[ ]:
!aws s3 cp edges.csv s3://$BUCKET/sm-model-lineage-export/
!aws s3 cp artifacts-nodes.csv s3://$BUCKET/sm-model-lineage-export/
!aws s3 cp tcs-nodes.csv s3://$BUCKET/sm-model-lineage-export/
!aws s3 cp action-nodes.csv s3://$BUCKET/sm-model-lineage-export/
!aws s3 cp nodes-contexts.csv s3://$BUCKET/sm-model-lineage-export/
Setup Neptune Cluster and notebook plugin
In the new few steps, we need to create a Neptune cluster either from the public CloudFormation template or manually from the console. After this cell is complete, restart the notebook’s kernel.
You can create the Neptune cluster from this CloudFormation template
Also, the instructions to install the Graph notebook jupyter extension to visualize the graph from Here. This is an Jupyter notebook extenstion to make it easy to communicate with Neptune cluster
Setup Neptune IAM Authentication Policy
Follow the instructions here: https://docs.aws.amazon.com/neptune/latest/userguide/iam-auth-enable.html
Change Graph config and point to Neptune Endpoint
If you get an error message: “UsageError: Cell magic %%graph_notebook_config not found.” use Jupyter Notebooks instead of jupyterlab and make sure that you have already installed the Graph extension from HERE
%%graph_notebook_config
{
"host": "localhost", ##Change this value to point to Neptune Cluster.
"port": 8182,
"auth_mode": "DEFAULT",
"iam_credentials_provider_type": "ROLE",
"load_from_s3_arn": "",
"ssl": false,
"aws_region": $REGION
}
Make sure the configuration for the Neptune cluster is correct
%graph_notebook_config
Bulk load data into Neptune
The next section of the code can be converted into code cells after adding the missing variables
%%bash
curl -X POST -H 'Content-Type: application/json' \
https://<Neptune Cluster Endpoint>:8182/loader -d '
{
"source" : "s3://$BUCKET/sm-model-lineage-export/",
"format" : "csv",
"iamRoleArn" : "arn:aws:iam::$ACCOUNT_ID:role/NeptuneLoadFromS3",
"region" : $REGION
}'
Run a couple of Queries
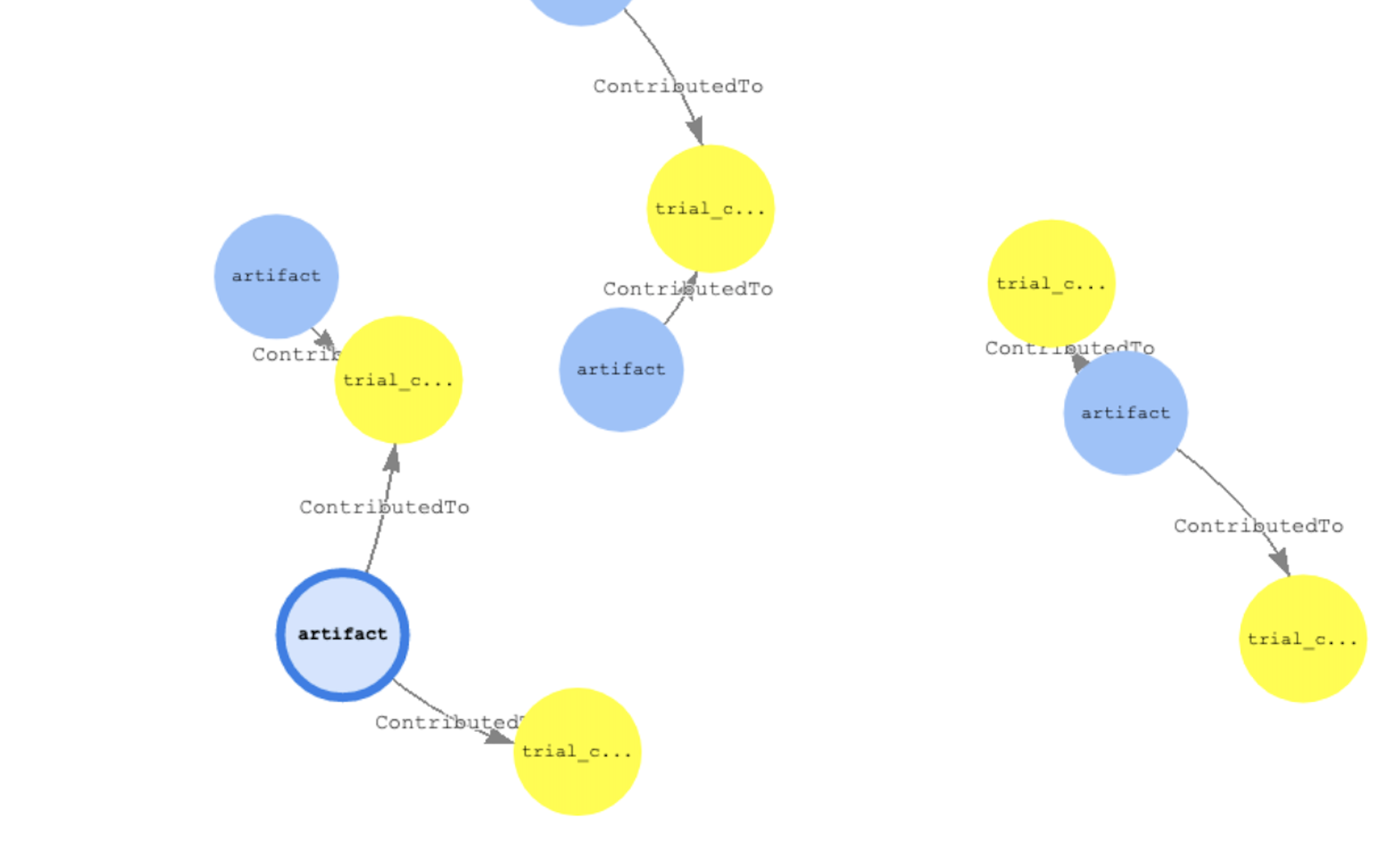
Let’s run a few queries to see some data lineage information. The first query will list all of the artifacts along with directional steps of outward edges and inward vectors
%%gremlin -p v,oute,inv,ine
g.V().hasLabel('artifact').outE().inV().path()
Here is an example of another query with more filters. We need to look at all the artifacts of type dataset then display some of its properties like: ARN, type, source ARN and status
%%gremlin -p v,oute,inv,ine
g.V().hasLabel('artifact').has('type','DataSet').outE().inV().path().
by(valueMap('arn','type','sourcearn','status'))
Clean up - Reset the Database
To clean up, you can reset and delete all of the data in the Graph database. This will involve 2 steps:
1- Initiate a database reset.
2- Execute database reset.
%%bash
curl -X POST \
-H 'Content-Type: application/json' \
https://<Neptune Cluster Endpoint>:8182/system \
-d '{ "action" : "initiateDatabaseReset" }'
%%bash
curl -X POST -H 'Content-Type: application/json' https://<Neptune Cluster Endpoint>:8182/system -d '
{
"action": "performDatabaseReset" ,
"token" : "<Token ID>" #Token ID is the reponse from the previous cell
}'
Notebook CI Test Results
This notebook was tested in multiple regions. The test results are as follows, except for us-west-2 which is shown at the top of the notebook.